F 1,6-BPase can be in one of two quaternary conformations, the R-state
(figures 4 & 5), which is catalytically active, and the T-state (figures
6 & 7), which is catalytically inactive. The binding of AMP triggers
the R-state to T-state transition of F 1,6-BPase causing a shift in the
binding sites for metals to an unfavorable position. The R and T forms
differ by a 17° rotation of the lower dimer C3–C4 relative to the upper
dimer C1–C2 and by a 1.9° rotation of the AMP binding domain relative
to the Fructose 1,6-bisphosphate domain (6).
Divalent metals like Mg2+,
Mn2+, and Zn 2+ are essential for F 1,6-BPase activity. Monovalent ions
like K+, Rb+, and NH3+ further enhance reaction rates at low concentrations
but can be inhibitory at high concentrations (7).
The reaction of F 1,6-BPase is pH-dependent. At neutral pH, plots of initial
reaction velocity versus Mg2+ are sigmoidal with Hill coefficient of 2
while at pH of 9.6 the plots are hyperbolic. The kinetic mechanism at pH
7 with Mg2+ as the cation activator is steady state random, whereas at
pH 9.6 the kinetic mechanism is rapid-equilibrium random (7). AMP and fructose
2,6-bisphosphate inhibit the activity of F 1,6-BPase, AMP inhibition is
allosteric while competitive inhibition occurs between fructose 2,6-bisphaosphate
and the substrate fructose 1,6-bisphosphate at the active site.
 |
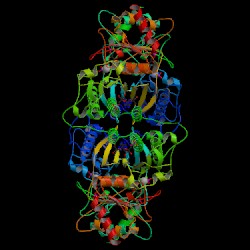 |
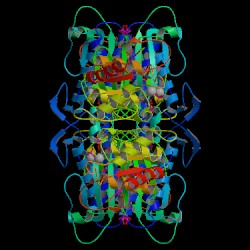
Figure 4. Crystal structure of
F 1,6-BPase complex with magnesium, fructose 6-phosphate and phosphate
(R-state).
Biological Molecule. Choe J., Honzatko
R.B. RCSB
Protein Data Bank. |
Figure 6. Crystal structure of
F 1,6-BPase complex with AMP, Magnesium, fructose 6-phosphate and phosphate
(T-state). Biological Molecule. Choe J., Honzatko R.B. RCSB
Protein Data Bank. |
 |
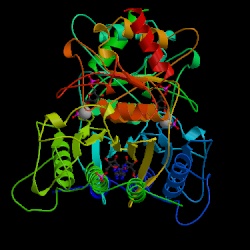 |
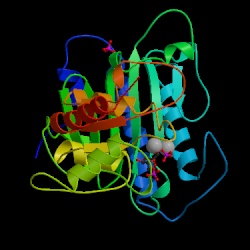
Figure 5. Crystal structure of
F 1,6-BPase complex with Magnesium, fructose 6-phosphate and phosphate
(R-state).
Asymmetric Unit. Choe J., Honzatko
R.B. RCSB
Protein Data Bank. |
Figure 7. F 1,6-BPase complex with
AMP, Magnesium, fructose 6-phosphate and phosphate (T-state). Asymmetric
Unit. Choe J., Honzatko R.B. RCSB
Protein Data Bank. |
|
|