|
1) Genetic
Architecture of Apomixis — My previous work has shown that apomixis in Erigeron is regulated by two separate
loci. Additional experiments have
explored the inheritance of these loci and demonstrated that apomictic
development can be recreated by experimentally recombining the two separate
gene regions. My pending proposal to
the National Science Foundation (January 2006 – Biology: Genes and Genome
Systems Cluster) is to develop two mapping populations (one based on maternal
segregation and the other based on paternal segregation) as robust sources of
markers linked to apomictic reproduction.
These markers will provide a means to explore the genetic and physical
structure of apomictic genome regions.
My ultimate questions relate to the origin and evolutionary history of
these regions.
|
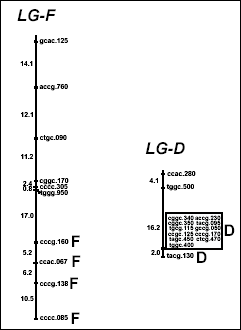
Fig.
1. Genetic model. Apomixis in Erigeron annuus is controlled
by two separate loci: ‘D’ causes
diplospory (unreduced gametophyte formation); ‘F’ induces autonomous embryo
and endosperm formation. These two
factors segregate independently in crosses to yield four phenotype classes.
|
|
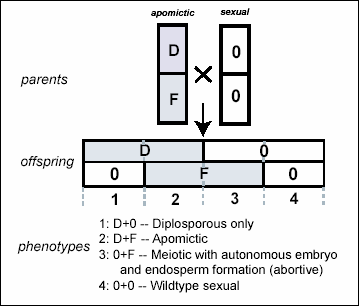
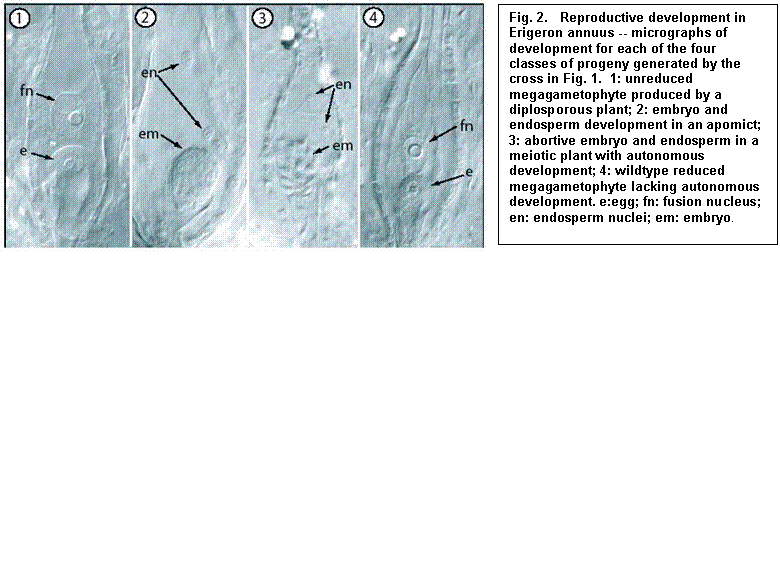
Fig.
3. Genetic map for
apomixis in Erigeron annuus featuring separate linkage groups for ‘D’ and ‘F’. Map distances between AFLP markers given
in centimorgans.
|
|