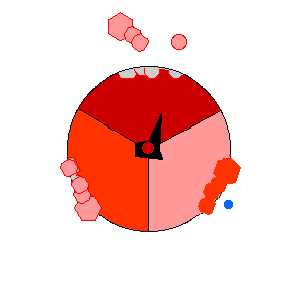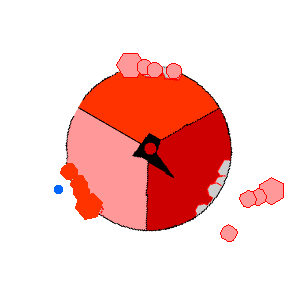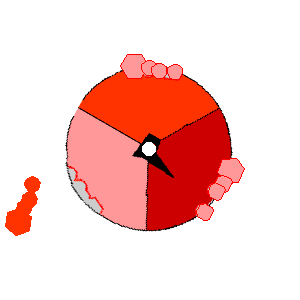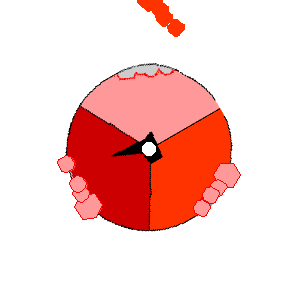
Image from http://rsb.info.nih.gov/NeuroChem/biomach/ATPsyn.html showing ATPase in action (open, loose and tight conformational states)
Mitochondria & Respiration
I. General Observations
1. Mitochondria are a cellular site for the synthesis of ATP
What type of cells contain mitochondria?
2. Microscopic observations
Distribution
II. Mitochondrial Structure
A. General features: two membranes/two compartments
2. cristae
3D rotation showing cristae inner membrane connection color coded reconstruction
tubular cristae from testes mitochondria (associated with steroid synthesis)
Were the "lollipops" just artifacts of EM preparation?
3. Matrix
III. Mitochondrial Fractionation
B. Example marker enzymes in the mitochondrion
| Inner membrane | Outer membrane | Matrix | Intermembrane compartment |
| 21% total protein | 6% total protein | 67% total protein | 6% total protein |
| respiratory chain enzymes, succinate dehydrogenase | monamine oxidase, fatty acid thiokinase | pyruvate dehydrogenase complex, citrate synthase, fumarase | adenylate kinase, nucleoside diphosphokinase |
IV. Molecular Organization and Function of the Mitochondria
A. Pathway of Biological Oxidation in Eukaryotes
starch, glycogen--> (glycolysis)--> pyruvate--> pyruvate
dehydrogenase complex--> acetyl CoA
triacylglycerol--> (lipase)--> fatty acids--> (fatty
acid oxidation)--> acetyl CoA
Features of mitochondrial oxidation
1. pyruvate, fatty acids and amino acids
2. acetyl CoA
3. NAD
and FAD
4. electron carriers in the inner membrane (ETS)
B. Isolation of the Components of the ETS
1. NADH dehydrogenase (16 - 26 polypeptides)
2. Cytochrome b - c1 (8 polypeptides)
3. Cytochrome oxidase (13 polypeptides)
4. ATPase coupling
factor (19 - 22 polypeptides)
C. Two issues to discuss
1. Energetics
Redox potential
What is redox potential? Give an example of a conjugate redox pair. A
conjugate redox pair with a negative potential (in volts) is more likely to be an electron
donor or an acceptor? Which has the more negative redox potential, NADH/NAD+ or
H2O/1/2O2?
Note that there are large potential drops associated with electron transport in the inner membrane
Where did the energy originally come from that is released from these
reactions?
2. Mechanism
Note that the orientation
of the three complexes, plus the ATPase coupling factor is essential to the proper
functioning of this system?
Why is this asymmetrical orientation a crucial feature to this system?
Where do the electrons that are passed between the complexes come from? Where
do these electrons eventually go?
3. Relationship between oxidative phosphorylation and the ETS
- the proton motive force (PMF).
-Calculation of free energy difference associated with PMF
For example, a typical mitochondria involved in ATP synthesis at 37C with a
membrane potential difference of -0.16 volts (this means more negative in the matrix than
in the intermembrane space because of the outward pumping of protons) and a matrix pH that
is 1 unit higher than the surrounding cytoplasm:
(delta)Goutward = 2.303 RT (pHinside - pHoutside) - nFEm
= (2.303)(8.314 X 10 -3)(273 + 37)(1) - (1)(96.5)(-0.16)
= +21.4 KJ/mole of protons
This means that the outward pumping of protons against an electropotential
gradient is endergonic and requires 21.4 KJ/mole; to bring protons back into the matrix
will be exergonic (-21.4 KJ/mole)
The free energy difference is useful to understand features of the
energetics of ATP synthesis:
The (delta)G for ATP formation depends on the concentration of ATP, ADP and
Pi in the matrix;
ATP + H2O<--> ADP + Pi
At equilibrium,
(delta)G0 = -RT ln Keq
= -RT ln [ADP][Pi]/[ATP]
(delta)G0 (ATP) = -34 KJ/mole (chemist's value!)
In the mitochondria, it takes 3 protons translocated from NADH to make 1 ATP;
that is, (delta)G = -64.5 KJ/mole (biologist's value)
So, if the available energy is greater than -64.5 KJ/mole, ATP will be made. The rate of ATP synthesis is dependent on the concentrations of ATP, ADP and Pi, plus the rate of proton translocation (the electrochemical gradient).

Image from http://rsb.info.nih.gov/NeuroChem/biomach/ATPsyn.html
showing ATPase in action (open, loose and tight conformational states)