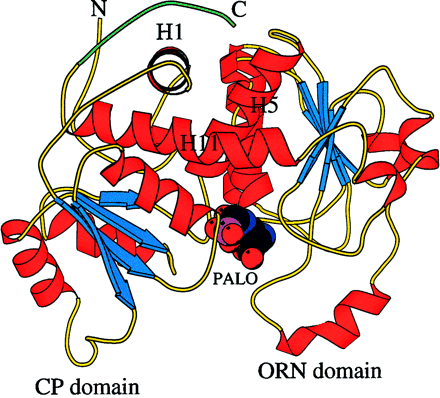
Fig. 2. Ribbon diagram of human OTCase liganded with the bisubstrate analog PALO. Alpha-Helices are shown in red, beta-strands in blue, and random coils in yellow. The C-terminal extension (residues 345-354), near helix H1, is shown in green. Helices H11 and H5 are inter-domain helices that link the CP and ORN domains. (6)
The CP binding domains are located in the interior of the protein,
and the binding domains for the ornithine are external. The interface between
the subunits is formed primarily by residues from the CP binding domains
of adjacent subunits. (6) The sequence of the C-terminal extension, found
only in mammals, is homologous to an interhelical loop found in several
membrane proteins, including mitochondrial transport proteins, which suggests
a possible mode of interaction with the inner mitochondrial membrane. (7)
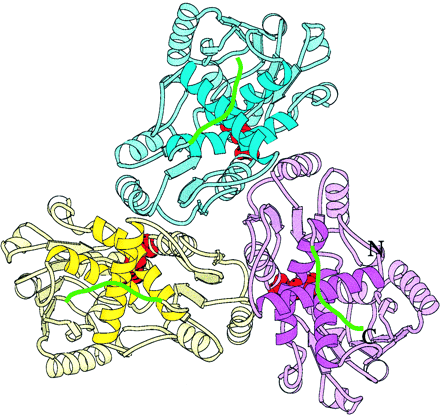
Fig. 3. Ribbon diagram of the human OTCase catalytic trimer.
The identical subunits are colored pink, blue, and yellow. The bisubstrate
analog PALO, shown in red, interacts with residues from two adjacent subunits.
Helices H1, H5, and H11 are shown in brighter colors. The C-terminal extensions,
shown in green, are exposed on the convex face of the enzyme. (6)
MECHANISM
OTC undergoes ordered, sequential displacement. Fist the CP binds
in a deep pocket, then the Orn closes up the active site above the CP.
(8) Next, the products are formed and released. The mechanism of
forming the citrulline and phosphorus products is as follows. The
sidechain of Cys303 is positioned so as to be able to interact with the
gamma-amino group of L-ornithine which attacks the carbonyl C of carbamoyl
phosphate in the enzyme-catalyzed reaction. This sulfhydryl group forms
a charge relay system with Asp263 and the alpha-amino group of L-ornithine.
(7) (See figure 4)
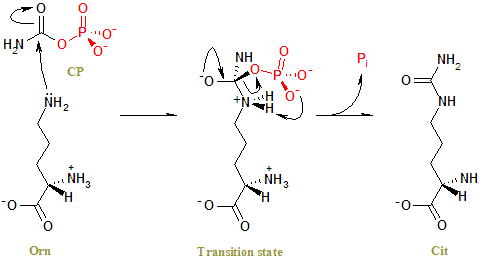
Fig 4. The reaction mechanism of OTC. The side chain amino group of Orn undergoes a nucleophilic attack on the carbonyl carbon of CP to form a tetrahedral transition state. Charge rearrangement releases Cit and Pi. (1)
The active site of OTC has been observed using the crystal structure
of OTC complexed with thebisubstrate analog PALO. (6) When compared
to the well documented and very similar active site activity of E.
coli ATC, OTC’s active site chemistry has been proposed. (12) PALO mimics
the bound CP and Orn, so the amino acid residues that interact with PALO
in the acive site are the same residues that interact with the normal substrates.
Figure 5 shows the interaction of PALO in the active site of OTC.
Ser-55 through Thr-58, Arg-106, Arg-319, and Gln-82 from the neighboring
subunit stabilize the negative charge of the phosphonate of PALO, so they
must interact with the phosphate on CP. The part of PALO that mimics
Orn is hydrogen bonded to the main chain carbonyl oxygen of Leu-274, to
the nitrogens of Arg-106 and Arg-319, and interacts with Ser-235 and Met-236.
Lys-53, Ser-55, Arg-57, Thr-58, Glu-87, Arg-106, His-133, Gln-136, Asn-167,
Asp-231, Ser-235, Met-236, Leu-274, and Arg-319 all interact with the bound
substrate analog also. (4) Some scientists were able to bind CP alone to
human OTC and found specifically which amino acids in the active site were
involved with its binding. Ser-90, hr-91, Arg-92, Thr-93, and
Arg-141 from one subunit and His-119 from the adjacent subunit are involved
in binding the phosphate group of CP. Gln-191, Cys-303, and
Arg-220 are involved in the interaction with the primary nitrogen of CP.
Thr-93, Arg-141, and, His-168 are involved in binding the carbonyl oxygen
of CP. (8) The available crystal structures clearly indicate that
the Asp-Xaa-Xaa-Xaa-Ser-Met-Gly sequence in human OTC from the flexible
SMG loop is the main binding site for ORN in OTC. (8) (see Figure 6)
Figure 6. Ribbon representation of the OTCase trimer. The three monomers
are coloured blue, red and yellow. Note the SMG loop. (3)